Biography
Hello! I am a research scientist with 20 years of deep experience in the fields of bioinformatics, genomics, and structural biology. I thrive when working on multi-disciplinary projects that combine genomics and computational (ML/AI) approaches to explore and solve complex scientific questions.
I received world-class PhD training in biophysics, made the leap to computational chemistry to work on drug discovery as a postdoc, and now work as a computational biologist and data scientist.
Mission
I aim to accelerate biomedical discovery through the application of cutting-edge computational methods and data visualization. My focus is on leveraging my experience in genomic data analysis to enable better decision-making in both research and clinical settings.
Leadership
As Director of the Bioinformatics Division at IIHG and Senior Scientific Advisor for FBB-Biomed, I lead cross-functional teams in indentifying and applying innovative bioinformatic solutions for complex biological problems. My proven track record includes over 60 scientific publications in top journals, accumulating more than 3000 total citations (h-index, 19).
I am also a Co-founder of the UI Spatial Transcriptomics Consortium and Co-Director of the Hawk-IDDRC Developmental Genomics Core.
Consulting
I help early-stage “startup” biotech companies working in the genomics and diagnostics space organize, analyze and interpret their genomic sequencing and assay data. I have 2+ years of experience consulting in this space. Along the way I have helped my clients file 2 major provisional patents creating significant value for founders and investors.
Member, International Society of Computational Biologists (ISCB) 2015-present
Member, Association of Biomolecular Research Facilities (ABRF) 2023-present
- Multiomic data integration
- Mentoring junior scientists
- Machine learning for biomarker discovery
PhD in Biophysics, 2009
The Johns Hopkins University
B.S. in Biochemistry (with Honors), 2003
University of Iowa
Skills
Experience
Responsibilities include:
- Leading and managing a team of 3 bioinformaticians
- Improving core revenue by adopting ‘agile’ project workflows
- Testing Director for CAP/CLIA KidneySeq targeted NGS panel
- Co-founding and running the UI 10X Visium Spatial Consortium
Responsibilities include:
- Setting up and managing AWS pipelines for data analysis
- Exploratory data analysis and QC
- Creating predictive models using machine-learning
Activities:
- Created the IIHG YouTube channel (18+ technical videos, >2500 subscribers)
- Collaborated on wide array of human and mouse genomics projects
- Published many co-author papers in high-impact journals
Activities:
- Worked as a lead computational biologist on discovery team for novel p97 inhibitors
- Validated binding predictions with labeled, high molecular weight NMR measurements
- Performed MD simulations of protein-protein complexes to study binding free energies
Certifications & Courses
Featured Publications
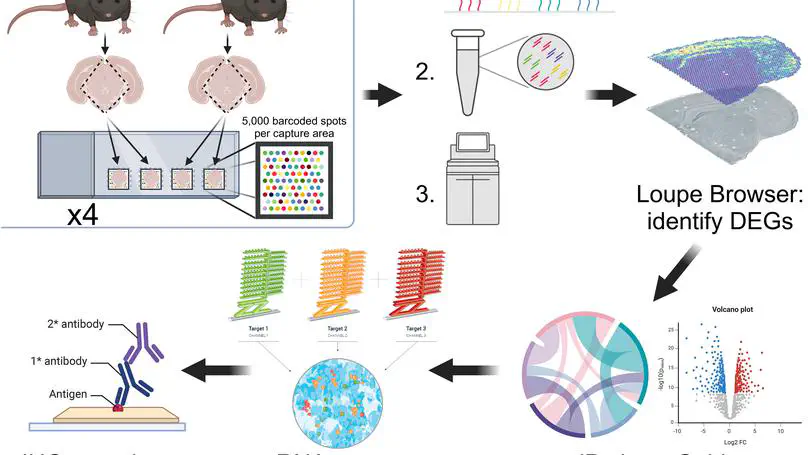
Map2k3os may serve regulatory roles relevant for tau phosphorylation and warrants further investigation to characterize its interactions. Overall, this report demonstrates the power of large-scale spatial transcriptomics technologies, while underscoring the need for convergent-data validation to overcome their limitations.
Recent Publications
Contact
Contact information and University of Iowa office location:
- michael dot chimenti at gmail dot com
- 319-335-6717 (University Office)
- 431 Newton Road, Iowa City, IA 52242
- Go to EMRB on the West Side of Campus and take the North elevator to the 3rd floor. My office is 329A.
